Opening up new opportunities for discovery
Research Applications:
- RNA-seq
- RT-PCR
- RT-qPCR
- Template Switching
- RNA Structure Probing
The process of reverse transcription (RT) enables researchers to “see” the transcriptome. However, most RT enzymes lack robust processivity - the ability to efficiently and continuously synthesize cDNA without dissociating from the RNA strand - making the transcription of long, highly structured, or modified RNA transcripts difficult. Furthermore, most RT enzymes operate at temperatures that can damage RNA, jeopardizing the detection of complex and low-abundance transcripts. As a result, researchers may be missing RNA transcripts in their samples.
UltraMarathonRT (uMRT) is a next-generation reverse transcriptase built to reveal transcriptomic diversity in biological samples. Structurally distinct from other RT enzymes, uMRT operates at ambient temperature, preserving RNA integrity. uMRT uses its unmatched helicase activity to unwind complex RNA structures and its enhanced processivity to fully transcribe long, structurally complex transcripts in a single pass and at single-cell levels, enabling you to capture transcript data that is more representative of the diversity in your sample.
Transcribe any RNA in a single pass
Transcribe entire >30kb viral genomes with enhanced enzyme processivity.
Operate at ambient temperatures
Preserve RNA integrity with lower reaction temperatures 30°C
Power through complex transcripts
Unmatched helicase activity unwinds complex RNA structure to enable complete 5’ end to 3’ end sequencing of previously invisible regions:
• Highly structured RNAs
• Centromeric regions
Detect RNA modifications
Uncover RNA modifications:
• Epitranscriptome
• RNA secondary structures
• RNA editing sites
• Post-transcriptional
Simplify library prep with efficient template switching
Create full-length cDNAs in a simplified reaction even when the 5' end of the RNA sequence is unknown
Learn more here
UltraMarathonRT kits are engineered to deliver more robust, unbiased data sets.
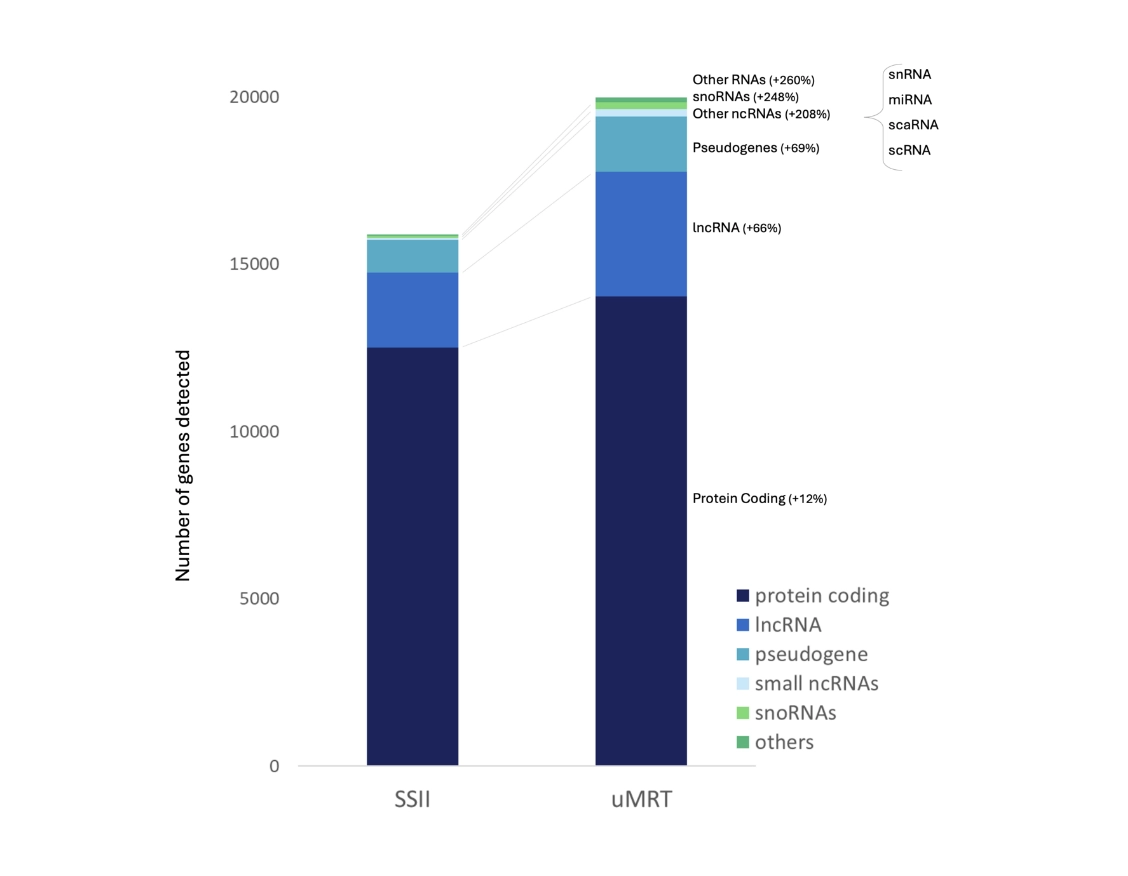
Identify more genes
Identify more genes
Changing the standard RT to uMRT enabled the identification of 25% more genes.
Comparison of uMRT and SSII in a representative SMART-seq experiment.
uMRT identified more genes in each RNA class.
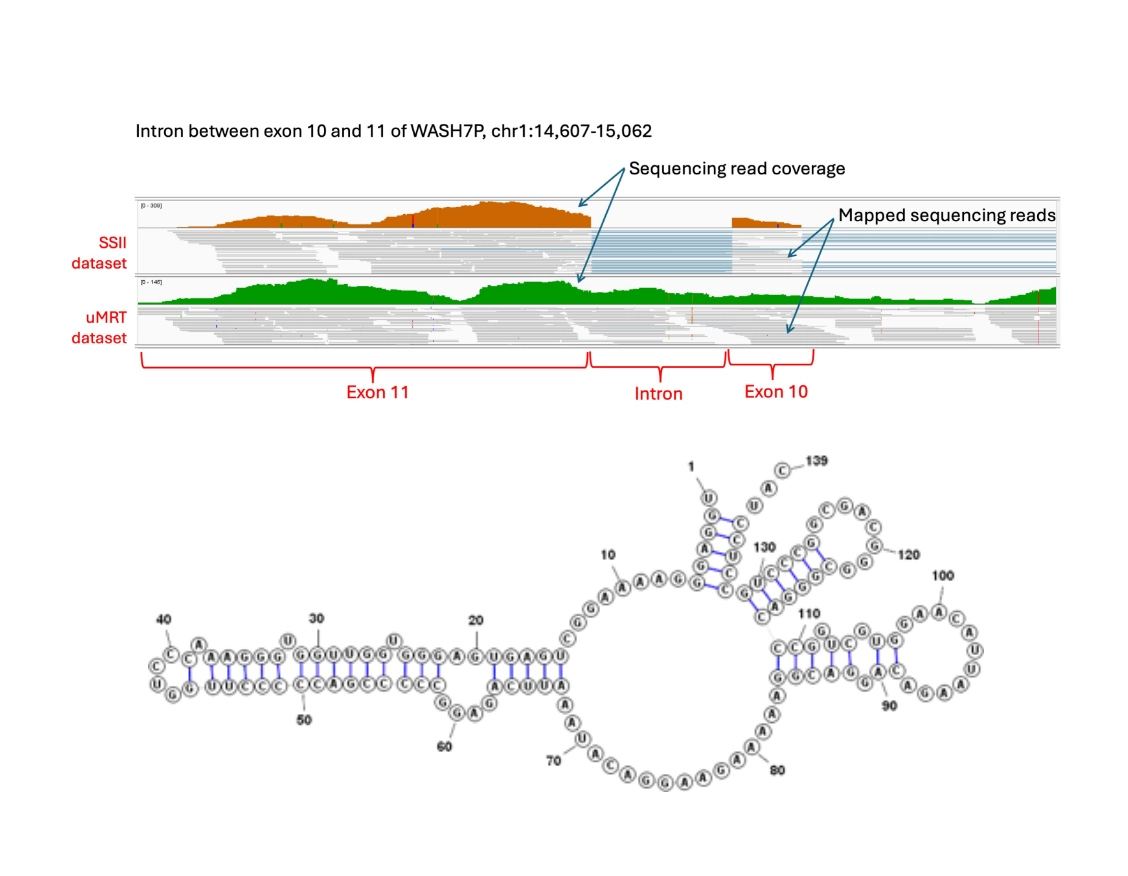
Discover challenging transcripts
Discover challenging transcripts
uMRT enables more accurate, unbiased transcriptome analysis
In a representative experiment, there were no sequencing reads from the SSII dataset mapped to the intron sequence between exon 10 and 11 of the WASH7P gene [top] which contains a stable structure [bottom].
In contrast, the intron can be efficiently detected by uMRT with its natural helicase activity.
Transcribe long RNAs
Transcribe long RNAs
uMRT shows strong performance with long transcripts
The UltraMarathonRT Two-Step RT-PCR Kit is ideal for applications requiring high sensitivity, particularly with low-abundance or structured RNA targets.
In the figure [right], amplification of transcripts are shown from 1.2 kb to 20 kb using 100 ng Hela total RNA. The cDNAs generated by Induro RT were amplified by LongAmp Taq. Reverse transcription was performed using Oligo(dT)18 primer, but the user can analyze multiple genes from a single RT reaction using a single RT primer. The power of UltraMarathonRT combined with an optimized DNA Polymerase provides superior performance.

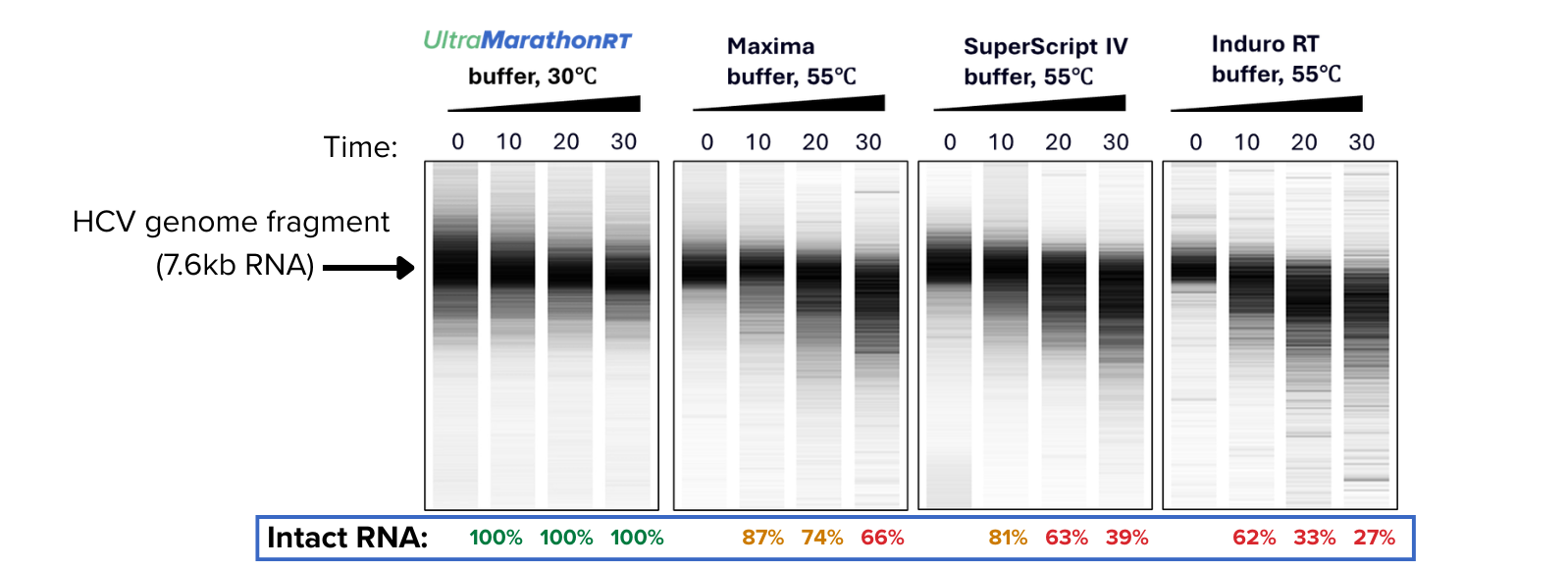
Protect RNA integrity
Protect RNA integrity
uMRT works optimally at ambient temperatures, preserving the integrity of RNA samples
RNA samples (particularly long, structured RNAs or very low abundant RNAs) are delicate and degrade rapidly when heated. uMRT works optimally at 20-42ºC.
In the figure [right], an HCV RNA was incubated in a diversity of buffer conditions without enzyme to evaluate the impact of temperature on template stability. Samples were analyzed using the Agilent 2100 Bioanalyzer system. Incubation in uMRT buffer at 30ºC shows no degradation of HCV RNA through 30 minutes.
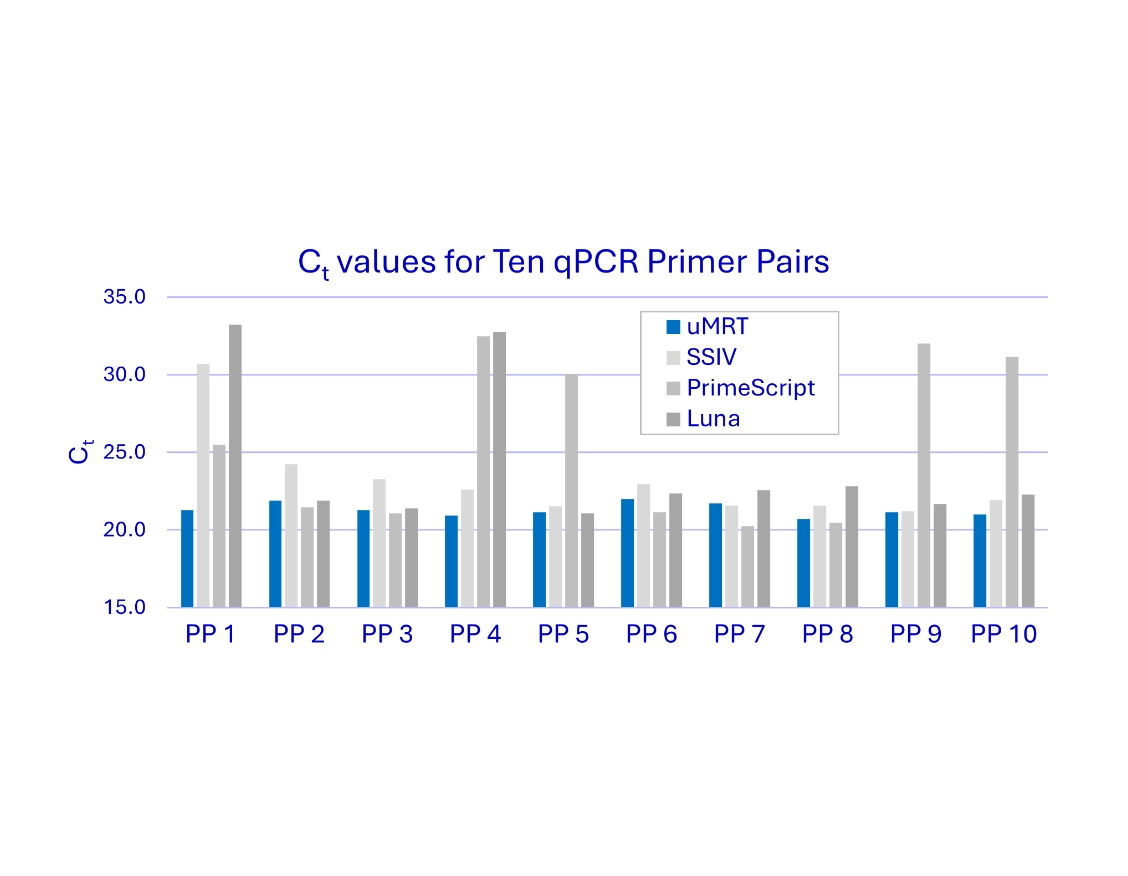
Simplify gene expression analysis
Simplify gene expression analysis
uMRT swiftly navigates high GC content and RNA secondary structures
The UltraMarathonRT® Two-Step RT-qPCR Kit sets a new standard in RNA quantification, delivering unmatched sensitivity, reliability,and robustness for researchers. In the figure [right], Ct values for four different RT-qPCR kitsacross all 10 primer pairs are shown. Because the uMRT kit shows no 5’ to 3’ bias and no bias for GC content, the results of the qPCR are very consistent. The competing kits have highly variable results
Identify more genes
Changing the standard RT to uMRT enabled the identification of 25% more genes.
Comparison of uMRT and SSII in a representative SMART-seq experiment.
uMRT identified more genes in each RNA class.

Discover challenging transcripts
uMRT enables more accurate, unbiased transcriptome analysis
In a representative experiment, there were no sequencing reads from the SSII dataset mapped to the intron sequence between exon 10 and 11 of the WASH7P gene [top] which contains a stable structure [bottom].
In contrast, the intron can be efficiently detected by uMRT with its natural helicase activity.

Transcribe long RNAs
uMRT shows strong performance with long transcripts
The UltraMarathonRT Two-Step RT-PCR Kit is ideal for applications requiring high sensitivity, particularly with low-abundance or structured RNA targets.
In the figure [right], amplification of transcripts are shown from 1.2 kb to 20 kb using 100 ng Hela total RNA. The cDNAs generated by Induro RT were amplified by LongAmp Taq. Reverse transcription was performed using Oligo(dT)18 primer, but the user can analyze multiple genes from a single RT reaction using a single RT primer. The power of UltraMarathonRT combined with an optimized DNA Polymerase provides superior performance.

Protect RNA integrity
uMRT works optimally at ambient temperatures, preserving the integrity of RNA samples
RNA samples (particularly long, structured RNAs or very low abundant RNAs) are delicate and degrade rapidly when heated. uMRT works optimally at 20-42ºC.
In the figure [right], an HCV RNA was incubated in a diversity of buffer conditions without enzyme to evaluate the impact of temperature on template stability. Samples were analyzed using the Agilent 2100 Bioanalyzer system. Incubation in uMRT buffer at 30ºC shows no degradation of HCV RNA through 30 minutes.

Simplify gene expression analysis
uMRT swiftly navigates high GC content and RNA secondary structures
The UltraMarathonRT® Two-Step RT-qPCR Kit sets a new standard in RNA quantification, delivering unmatched sensitivity, reliability,and robustness for researchers. In the figure [right], Ct values for four different RT-qPCR kitsacross all 10 primer pairs are shown. Because the uMRT kit shows no 5’ to 3’ bias and no bias for GC content, the results of the qPCR are very consistent. The competing kits have highly variable results

Research Applications:
Featured Applications
Click on the images to download the brochures
Our easy-to-use uMRT kits can be used to reveal more of the transcriptome, empowering discovery and translational science.