Perfect for researchers working with low abundance RNA, viral RNA, non-coding RNAs, long RNAs, and even highly structured RNA templates who demand enhanced gene expression analysis via RT-qPCR.
Unmatched Sensitivity: Enhancing and Streamlining Gene Expression Analysis
Key Features
Data Reliability
Provides consistent Ct value regardless of primer type (e.g. gene specific, oligo(dT) or randomer) or location. Other RT-qPCR kits will provide widely different Ct values if primers are in high vs low GC content areas, whereas the UltraMarathonRT® Two-Step RT-qPCR Kit will provide consistent Ct values regardless of location.
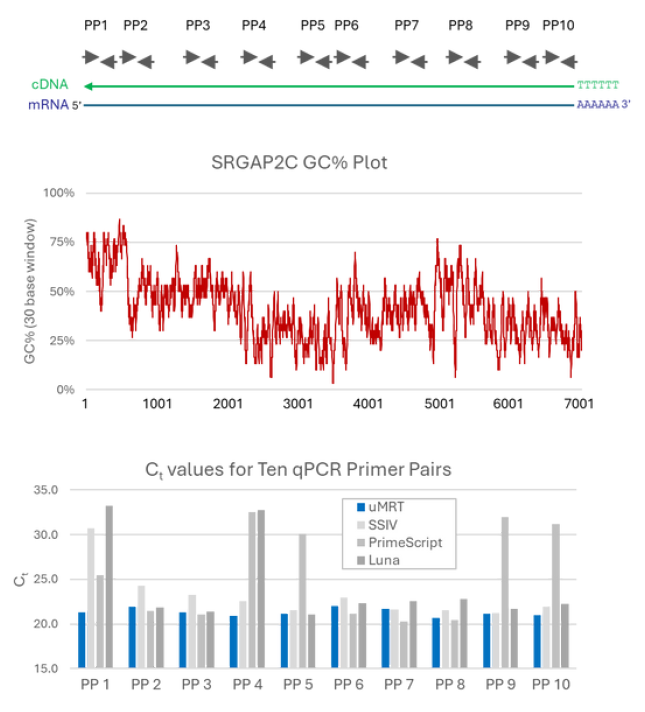
Achieve exceptional sensitivity and efficiency, ideal for detecting low abundance RNA, viral RNA, non-coding RNAs, long RNAs, and even highly structured RNAs. This makes it a versatile tool for a wide range of molecular biology applications.
Robust Performance
High Inhibitor Tolerance
Consistent Ct values are maintained in the presence of inhibitory compounds potentially found in biological sample preparation such as ethanol (resistant to 10%), isopropanol (resistant to 5%), TRIzol (resistant to 0.7%), Formalin (resistant to 0.0025%), Ammonium Acetate (resistant to 50mM), Bile Salts (resistant to 0.05%), and Hematin (resistant to 10µM).

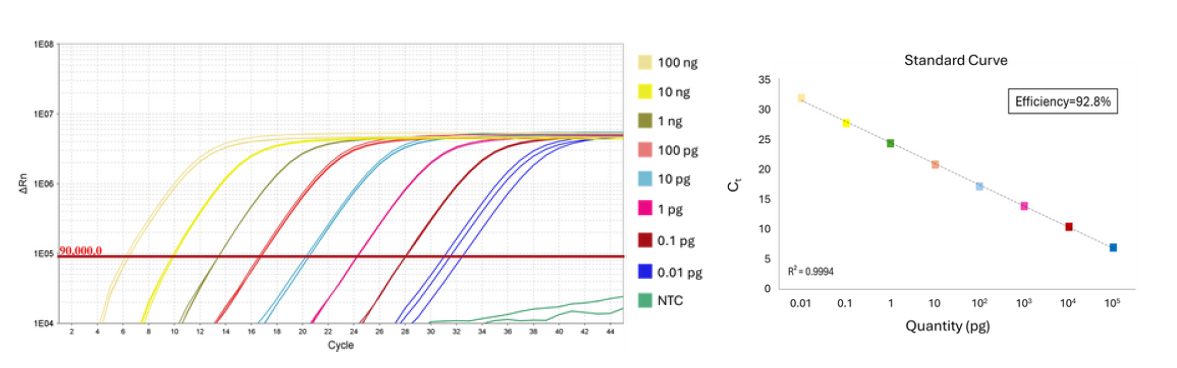
Capable of detecting RNA across a wide spectrum of input concentrations ranging from 0.1 pg to 2 µg, providing unparalleled flexibility in experimental design.
Broad Dynamic Range
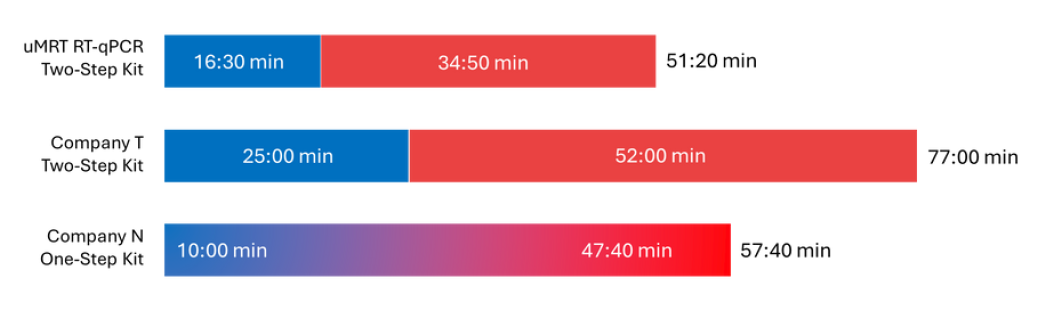
Minimal Experimental Time
An optimized protocol allows researchers to generate complete RT-qPCR data in under 1 hour.
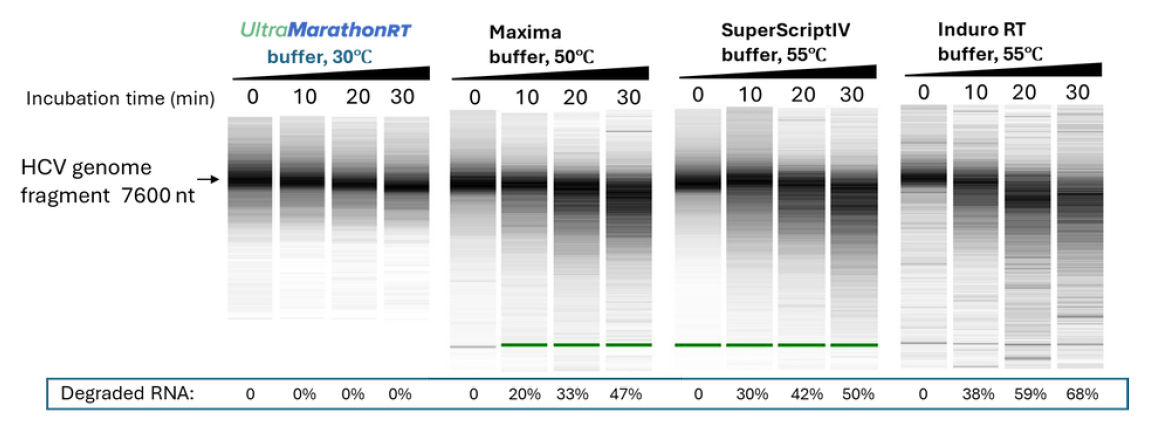
UltraMarathonRT has intrinsic helicase activity for RNA structure unwinding, making the high temperature reaction conditions utilized by other RTs totally unnecessary.
Preserves RNA Template Integrity
Dye-Based Detection
Incorporates reliable dye-based technology to facilitate accurate and reproducible real-time monitoring of RNA levels.
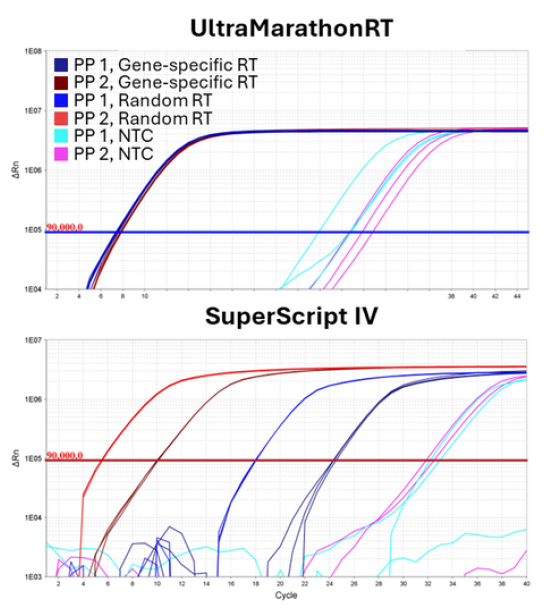
Only the UltraMarathonRT® Two-Step RT-qPCR Kit can reliably validate novel genes detected in uMRT RNA-seq data sets (genes that are undetected by other RTs).
Ideal Companion for uMRT RNA-seq Validation